To all methods, the upper capacity bounds apply as the edge multiplicities and, for the methods dealing with restricted node capacities, the node demands are interpreted as the node capacities.
The derived cuts a exported by the node colour register with the following interpretation:
For the edge connectivity and the strong edge connectivity case, the library includes methods to partition the node set such that all partial induced subgraphs are maximally connected. These methods formally return whether the graph object is connected and, if not, export the connected components by the node colour register. The respective codes for 1-edge connectivity, 2-edge connectivity and strong 1-edge connectivity run in linear time. The codes for high connectivity orders iterate on the min-cut methods, and hence are polynomial but not really efficient.
Concerning vertex connectivity, a partition into edge disjoint blocks is possible (and implemented) only in the low order cases: For 1-connectivity and strong 1-connectivity, the respective edge connectivity methods apply. The depth-first search method abstractMixedGraph::CutNodes() returns the 2-blocks (the edge-disjoint, maximal biconnected subgraphs) by the edge colour register, and the cut nodes by the node colour register. This procedure
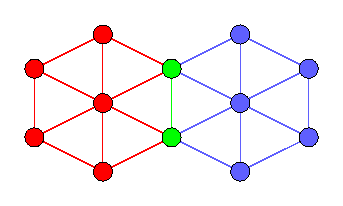
Starting with k=3, the maximal k-connected subgraphs (called k-blocks) are in general not edge disjoint as the following graph illustrates:

Herein, the green nodes form a cutting pair, and the green edge is contained in both 3-blocks. There is the concept of SPQR-trees to represent the 3-blocks, but this is out of the library scope.
Here are some caveats:
Although the library code is very elaborate, there are also theoretical limitations: The planar biconnectivity augmentation problem is NP-hard. For the planar triconnectivity augmentation of biconnected graphs, it is not even known if a polynomial time method is possible.
Dropping the planarity requirement admits linear time augmentation methods, but no such library code is available.